Useful Scripts and Tools#
Utility Codes Repo#
The NRLMOL_utility_codes GitHub repository has several useful tools:
- wfout_reader.f90:
Convert a binary WFOUT into a human readable format WFOUT2
- wfout_unp2pol_converter.f90:
Convert a spin unpolarized WFOUT into a spin polarized WFOUT.
- evalueSmearing.f90:
Apply Gaussain smearing on eigenvalues for plotting with gnuplot.
- xmol2cluster.pl:
Converts an xmol geometry file to NRLMOL CLUSTER file.
- qchem2cluster.pl:
Converts an qchem geometry file to NRLMOL CLUSTER file.
- cluster2xyz.pl:
Converts NRLMOL CLUSTER file to xyz file.
- benchmark_libxc.sh:
shell script testing suite code for the UTEP-NRLMOL code (written for old versions around 2015-2017).
- nrlmol2molden.f90:
Converts NRLMOL output to molden file
Visualization Tools#
Atomic Simulation Environment (ASE)
This easy to use visualization helps with quick editing and visualization that can be executed from the command line.
ase gui file.xyz
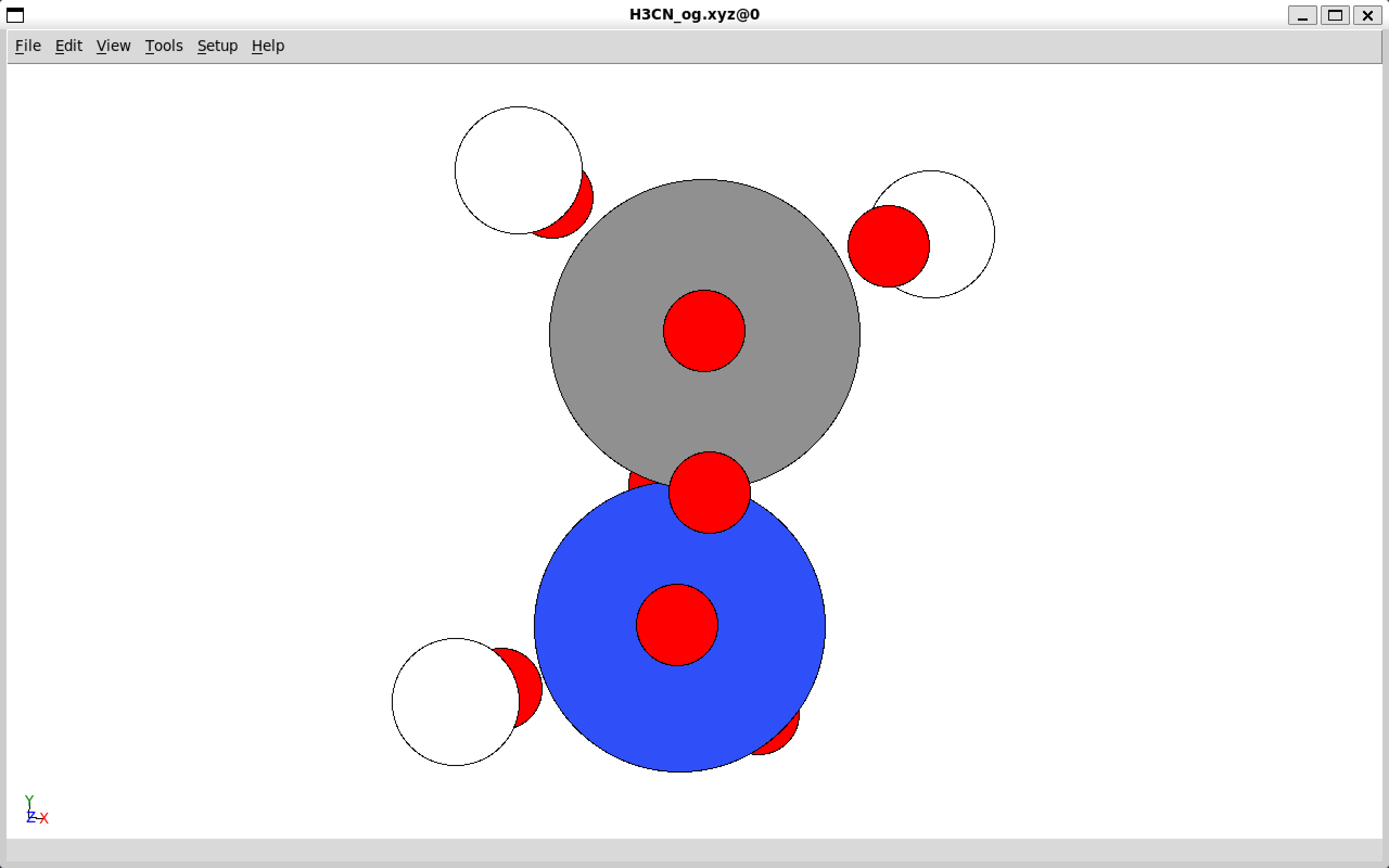
Sample output of visualizing an .xyz file with ASE.#
Visual Molecular Dynamics (VMD)
This software contains logical visualization, where you can create an image of only atoms of a certain category or type. You can also easily superimpose alternative views of a system (e.g. display ribbons and balls/sticks at once), and easily colorize the
With the atomic blender extension for the Blender software, you can create very high quality visuals. The only downside is that Blender has a high learning curve, but you can see a few examples of visuals done in Blender.
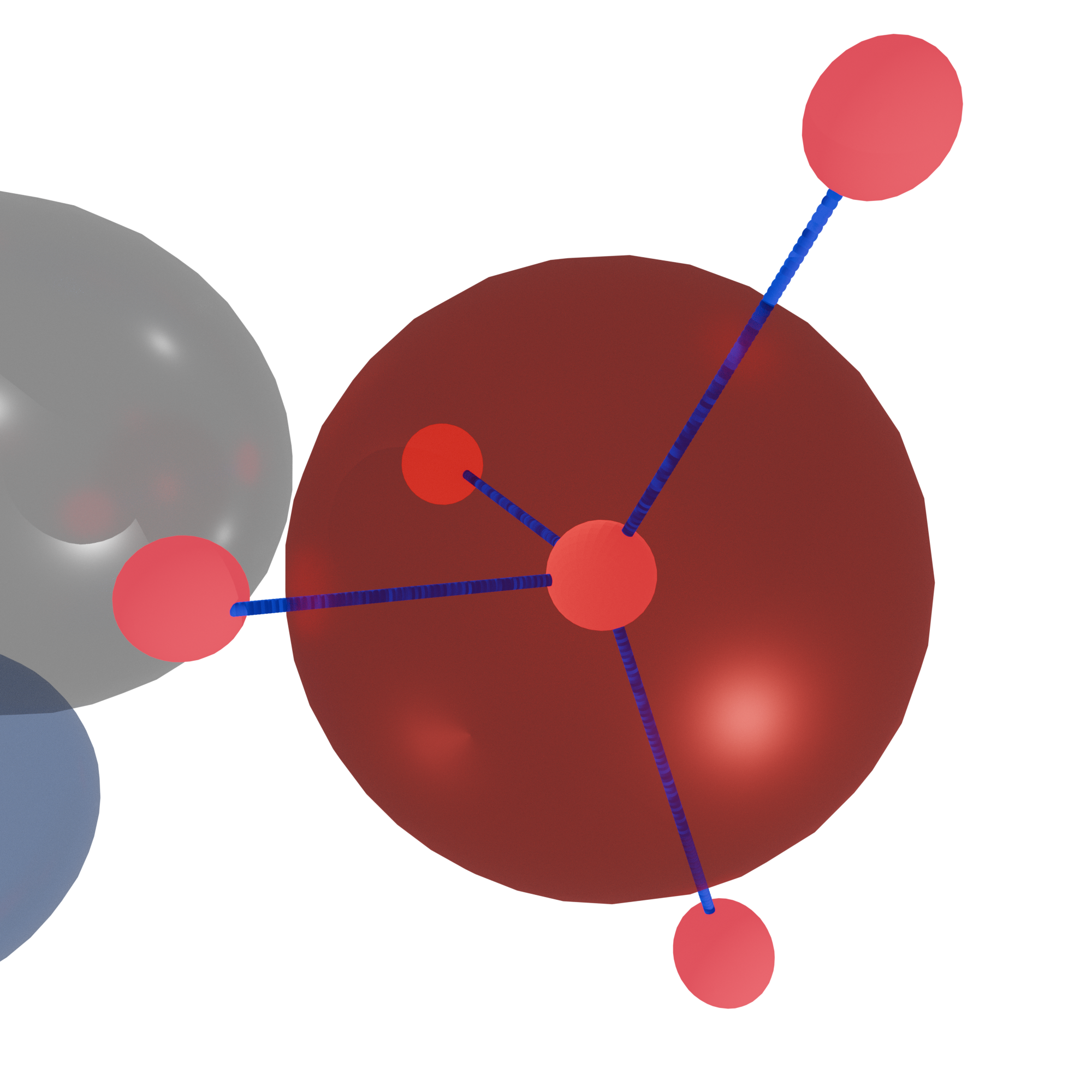
This figure shows how FODs create tetrahedra, or pyramids, in their SP3 form.#
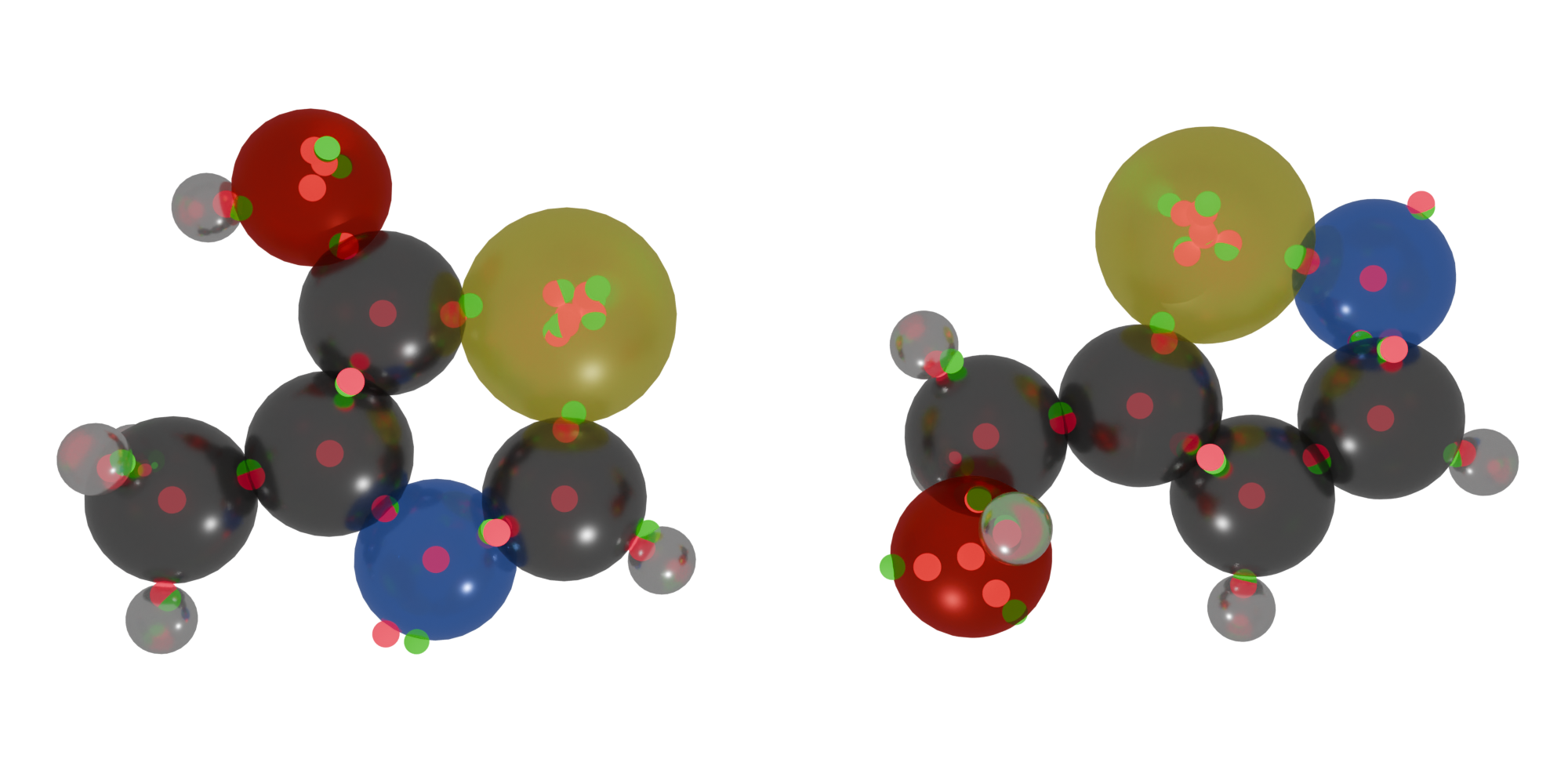
Sample isomers with FODs, rendered with Blender.#
Blender Learning
You need to learn how to manipulate cameras in Blender and how to do basic object manipulation, lighting, mesh editing, and texture editing. The first 11 videos in this tutorial series offers guidance in these basic things. Then you can use the extension!